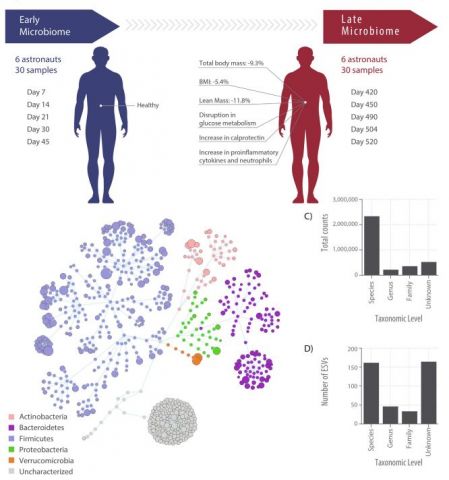
Deep space exploration is the future of spaceflight, but for this to be feasible, maintaining astronaut health is essential for long-duration spaceflights. The Mars500 experiment, in which a human isolation study was conducted over a 520-day period, analyzed fecal samples during the early and late stages of the experiment. A recent publication by Drs Nicholas Brereton and Emmanuel Gonzalez, “Reanalysis of the Mars500 experiment reveals common gut microbiome alterations in astronauts induced by long-duration confinement,” details how reanalyzed data from the Mars500 experiment (GLDS-191) lead to discoveries related to astronaut health. GeneLab recently spoke to Dr Brereton, from University of Montreal, and Dr Gonzalez, from McGill University, about their work and this interview highlights how the GeneLab Database enabled this reanalysis.
GeneLab: What were the most important findings in this study?
Nicholas Brereton and Emmanuel Gonzalez: The Mars500 experiment was the longest controlled human confinement experiment ever conducted, isolating six crew members in a ground-based simulated Mars habitat for 520 days in order to test the physiological and psychological impact of long duration space missions. The crew were largely healthy after 520 days but did have uniform muscle mass and strength loss, disrupted nutrient metabolism (fasting glucose was at prediabetic levels) and increased markers for intestinal inflammation. This reanalysis study explored the potential for common gut microbiome shifts in the crew throughout this isolation which could potentially be associated with these observed symptoms.
Using sequence data provided by GeneLab, significant shared changes to the relative abundance of microbiota were detected in the crew after 520 days. These included depletion of species of bacteria involved in degradation of resistant starch, nutrient absorption, and prevention of intestinal inflammation within the healthy human gut, such as Faecalibacterium prausnitzii, Ruminococcus bromii, Blautia luti, Anaerostipes hadrus, Roseburia faecis, and Lactobacillus rogosae. While these reductions in beneficial bacteria make sense with disruption of crew metabolism, significant increases were also identified in unknown “yet-to-be-cultured” bacteria after 520 days, which are still an important mystery left to be solved.
The discovery of significant alterations to specific microbiome species after long-duration confinement represents an important clue linking the physiological changes observed across different studies in the Mars500 crew and reveals the type of health challenges which astronauts might face during extended space flight.
GL: How did the GeneLab database and tools assist this work?
NB and EG: This research was funded by the Canadian Space Agency with the aim of mining and reanalyzing the valuable genomic data generated from experiments within space science and is the second time we’ve used GeneLab to identify a study where reanalysis was highly likely to generate novel discoveries. The care in which data deposited in GeneLab is curated, particularly the detail to which raw sample data is carefully linked to experimental factors and metadata, was essential for us to effectively explore and compare the experimental designs and methodologies used across different experiments. This allows the identification of studies where novel biological questions can be posed by simply altering experimental design or where analysis can be improved using new technologies and opens up an extraordinary number of opportunities for biological discovery within existing datasets.
GL: What are the next steps?
NB and EG: Improved understanding of the common microbiome alterations could help inform strategies to safeguard astronaut musculoskeletal and metabolic health during long-duration spaceflight, such as a manned mission to Mars.
One of the other aspects of this research was a direct comparison between data from the crew gut microbiota to a second environmental microbiome dataset from the sealed habitat’s surfaces. By comparing these, it was possible to gain a unique perspective into the extreme extent to which the human and environmental microbiome ecosystems interact during confinement. We hope to further develop technologies and strategies which enable comparisons between experiments and microbiome datasets to better reveal astronaut-environment microbiome interactions but also, more generally, to improve upon our understanding of the microbial dialog between humans and their habitats.

