A collaboration between NASA GeneLab and the Gilroy Lab has led to a multi-omics, cross-species analysis of GeneLab omics data, creating a new NASA Space Biology sponsored investigation into the effects of spaceflight and microgravity simulations on plant-microbe interactions.
A suite of visualization tools to explore relationships between studies within the GeneLab data repository has been created by Richard Barker, a member of the Gilroy Lab at the University of Wisconsin, Madison, and co-lead of the GeneLab Plant Analysis Working Group (AWG). These apps aggregate diverse data types and allow users to find associations between the distinct GeneLab accessions. This has allowed the identification of new correlations in the data leading to the generation of novel hypotheses and the creation of protocols for experimental validation.
The visualization toolset includes the following:
- X-Species Transcriptional Explorer - iterative filtering tool for processed transcriptional data in GeneLab including: human, mouse, rat, fly, worm, yeast, rice, tomato, Arabidopsis, Brassica and Brachypodium.
- GeneLab X-Species Metadata – API-linked, GeneLab meta-data visualization that allows for the identification of links between studies with related meta-data fields.
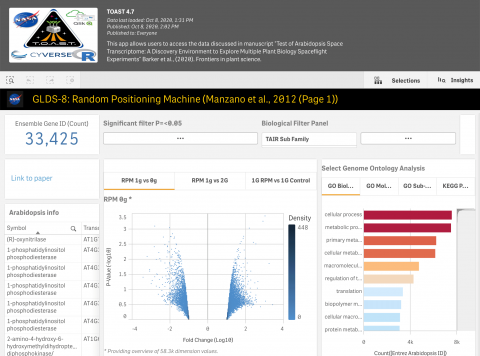
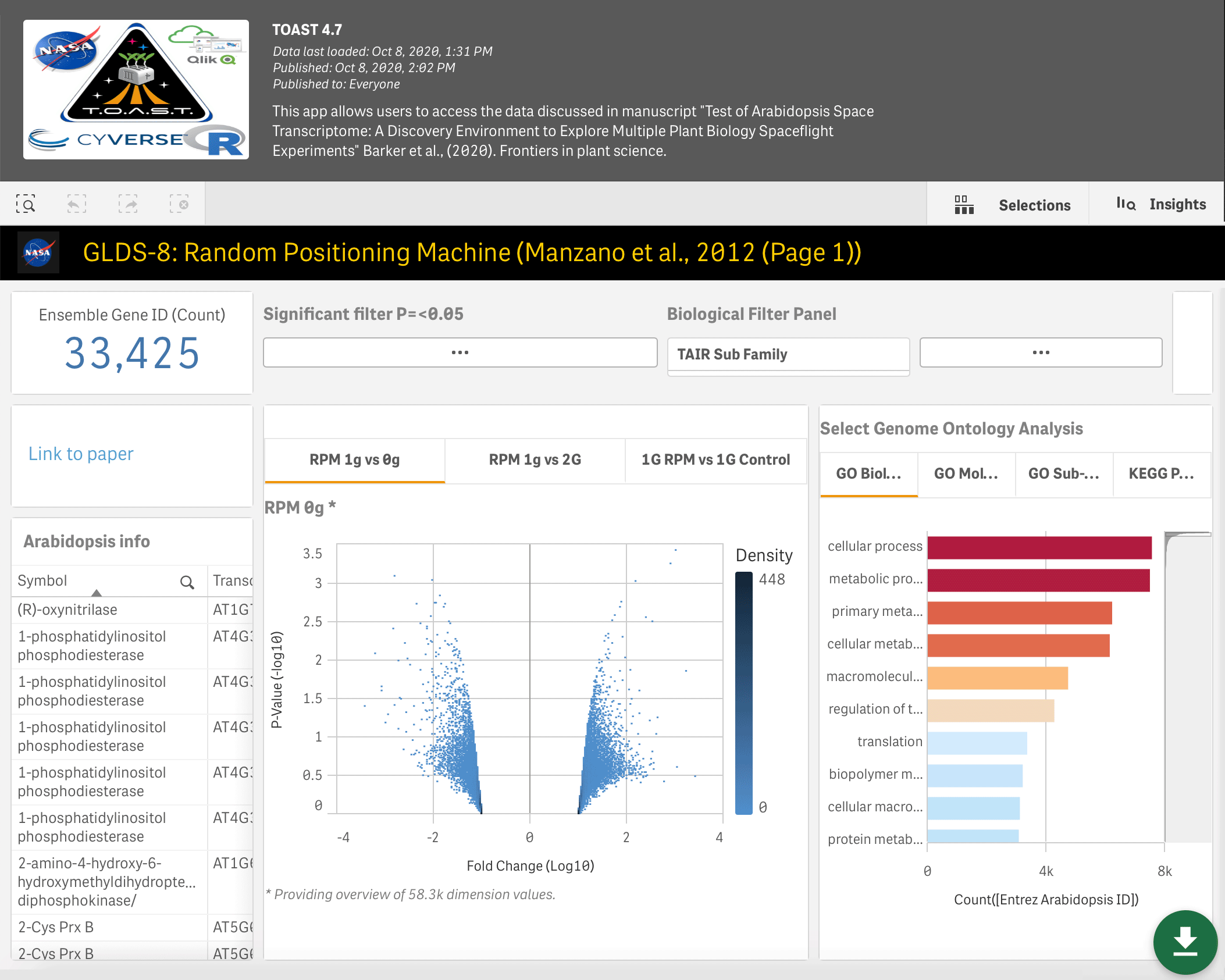
- TOAST - The AstroBotany multi-plant species and multi-omics explorer featuring: four types of microarray, RNAseq, epigenetics and proteomic data (Barker et al., 2020).
- MANGO ISS MicroBiome - Explorer for the ISS microbiome (GeneLab datasets GLDS-69 and GLDS-200). Microbial Observatory: indoor microbiome study of the ISS surfaces and the ISS dust particle microbiome (unpublished app).
- Plant-specific Metadata - To assist the GeneLab Plant AWG, a plant sample meta-data exploration system that allows users to find links between studies.
- Rad Space Bio App - Explorer for space radiation exposure experienced by each GeneLab experiment (currently under peer review).
These data-mining tools were used by Simon Gilroy, Professor of Botany, Gilroy Lab, University of Wisconsin, Madison, to identify links between oxidative stress in spaceflight and potential effects on plant-microbe interactions. These responses have potential consequences for both pathogen response and symbiotic interactions during spaceflight. This work is an example of how the GeneLab system can be used to make new discoveries that influence future experimental design, with the goal of generating new insights into the biological response to spaceflight.
The Gilroy Lab also discovered a genetic pattern in GeneLab dataset GLDS-37, which identified a Reactive Oxygen Species signature associated with the plant Arabidopsis thaliana in spaceflight (Choi et al., 2019). The data-mining tool TOAST allowed scientists in the lab to look for these and other transcriptional responses across multiple studies in GeneLab. TOAST analysis confirmed a strong link between oxidative stress responses and spaceflight but also expanded these flight-related signatures to include genes related to the TCA cycle in the mitochondria (Barker et al., 2020, TOAST GeneLab mining tool). These analyses provided a set of marker genes characteristic of plant spaceflight responses.
The Gilroy Lab built the GeneLab X-Species Transcriptional Explorer app to perform comparative transcriptomics with crop species such as rice and tomatoes. This approach allowed them to identify similar genes (i.e., orthologs) in these crop species to those that change in spaceflight in Arabidopsis. By mining the literature it was found that some of these orthologs were also known to be induced in tomatoes in response to beneficial microbes on Earth. It’s conceivable that these results could be pointing at possible gene expression changes in plants in spaceflight that could alter or mimic plant-microbe interactions.
The MANGO tool was employed to identify microbes that have already been detected on the ISS and reported to trigger or suppress the expression of the spaceflight marker genes in crop plants on Earth. This led to the identification of potential beneficial microbes that may alter the gene expression patterns seen in plants in spaceflight. The ability to identify the crop orthologs using the GeneLab X-Species Transcriptional Explorer meant that these questions of spaceflight-related, plant-microbe responses could be shifted from the model plant Arabidopsis to crop plants such as tomato.
With the award of the new NASA Space Biology solicitation, the Gilroy Lab will investigate this plant-microbe symbiosis on the ISS and ask whether interaction with these microbes alters tomato plant growth in space. The project will also focus on characterizing plant-microbe interactions in spaceflight analogs such as clinostats. Leveraging the clinostat data already deposited in GeneLab has helped design tests for how growth on the 3D clinostat affects both plant-microbe interactions and also how well this treatment mimics true spaceflight.

