
Data and metadata should be Findable, Accessible, Interoperable, and Reusable (FAIR). Following the FAIR guidelines, GeneLab has integrated a digital object identifier (DOI) with every dataset making it findable, accessible and reusable.
A DOI is a persistent identifier or handle used to identify objects uniquely, as standardized by the International Organization for Standardization. DOIs have become widely used in the sciences to identify publications of all kinds but primarily documents and data, and DOI registries now offer indexing and search functions across registered objects (e.g., DataCite is a widely used registry with search services for the sciences). NASA recently launched the Digital Object Identifier Management System (DOIMS) to help its customers use these registries to create and manage DOIs for their systems.
NASA GeneLab has integrated the data publication systems of its data repository - the first comprehensive space-related omics database - with the DOI management services provided by the DOIMS. Leveraging the Application Programming Interface or API, GeneLab data curators can now create DOIs for datasets contained in the repository, and associate these DOIs with valuable metadata to support the data search services provided by DataCite and other systems.
Once created, the DOI, along with guidance for citing the data by the DOI, are provided on each dataset’s webpage in the GeneLab data repository. Researchers can download this data citation and reference the data in publications and articles. In addition, GeneLab datasets are now searchable on DataCite (https://datacite.org/).
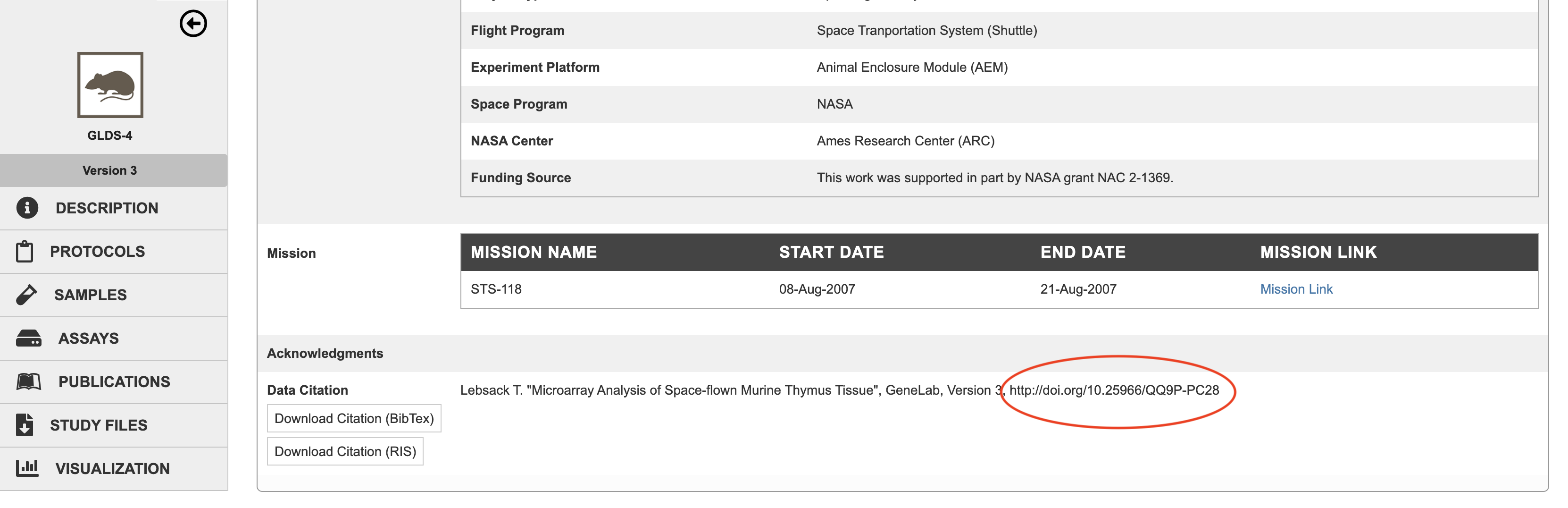
GeneLab dataset GLDS-4 illustrates where the DOI can be found within the dataset.
To learn more about FAIRness of the GeneLab Data Systems, check out the data versioning feature and FAIRness and Usability for Open-access Omics Data Systems.

