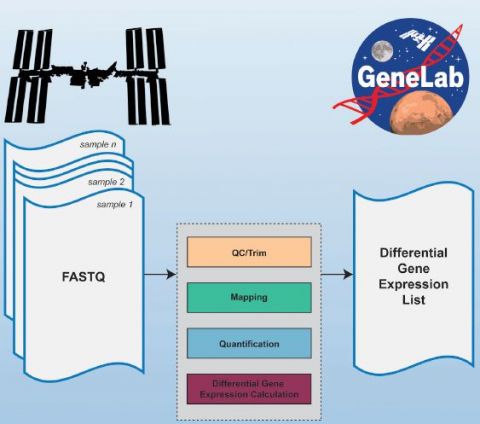
Utilizing technologies that allow for the greater understanding of how spaceflight affects human biology is essential to support deep space exploration. One such technology, called RNA sequencing, allows for the quantification of precise changes in gene expression profiles from organisms exposed to spaceflight. A recent publication, “NASA GeneLab RNA-seq consensus pipeline: Standardized processing of short-read RNA-seq data,” led by Drs. Eliah Overbey and Amanda Saravia-Butler, including contributions by 43 additional GeneLab Analysis Working Group (AWG) members, details the construction of a pipeline along with the rationale in an effort to promote transparency, reproducibility, and reusability of RNA sequence (RNA-seq) data. Dr. Overbey, from the University of Washington, and Dr. Saravia-Butler, from NASA’s GeneLab, are both contributing members of the GeneLab AWG. Recently GeneLab spoke to the authors about their work, and this interview highlights how the GeneLab AWG’s enabled the creation of this pipeline.
GeneLab: What was the motivation behind writing this publication?
Eliah Overbey and Amanda Saravia-Butler: The motivation behind developing and publicizing the GeneLab RNA-seq consensus pipeline (RCP) is to democratize space-relevant RNA sequence (RNA-seq) data hosted on GeneLab while minimizing bias. Raw RNA-seq data are only interpretable by a small subset of scientists known as bioinformaticians whereas differential gene expression data (DGE), an indication of which genes are turned on and turned off in response to the space environment, are accessible to a much broader scientific community. There are several tools and pipelines that can be used to generate DGE data, so to minimize bias GeneLab worked with the scientific community through our Analysis Working Group (AWG) members to develop an RCP. The RCP was designed to generate DGE data for the broad range of model organisms used in space-relevant studies hosted on GeneLab. Since each set of DGE data was generated with the same pipeline, there is no bias due to using different data processing tools when comparing these data across different studies. We decided to write this publication to ensure that all GeneLab users understand the rationale behind the RCP as well as all the processed data generated from the RCP that is made publicly available on the GeneLab Data Repository.
GL: How different is the GeneLab RNA-seq pipeline from other RNA-seq pipelines?
EO and ASB: The GeneLab RCP uses publicly available tools that are common to other RNA-seq pipelines, however the specific selection of the tools and parameters used is unique. The initial pipeline draft was first derived at a GeneLab workshop in Orlando, FL in April 2018. We started by identifying the tools popular among the bioinformatics-savvy users of the GeneLab database. Over the next year, we refined the specific tools and parameters further until reaching a consensus pipeline among GeneLab’s AWG members.
GL: What is the importance of using biological vs technical replicates and how many biological replicates are necessary for an RNA-seq experiment?
EO and ASB: Biological replicates are incredibly important in all biology experiments, and RNA-seq experiments are no exception. The more biological replicates in an experiment, the more confident scientists can be in the results, so we recommend as many biological replicates as the person conducting the experiment can afford with a minimum of 4 biological replicates per group for RNA-seq experiments. In RNA-seq experiments, technical replicates are primarily used to ensure consistency when generating libraries, sequencing samples in different lanes of a flow cell, different flow cells, or different sequencing instruments. When possible, I recommend all samples from a single study to undergo library preparation by the same person on the same day, and to be sequenced on the same lane(s) of a flow cell and sequenced at the same time on the same flow cell using the same instrument to minimize instrument bias. When possible, it’s better to include more biological replicates than technical replicates in the experiment.
GL: How did the GeneLab Analysis Working Groups help you achieve your goals?
EO and ASB: The GeneLab Analysis Working Group members were instrumental in developing the RCP. The pipeline framework was developed during the first Analysis Working Group workshop in Florida in 2018 and was continually refined with input from the Analysis Working Group in subsequent years. Their bioinformatics expertise as well as their vast experience in working with RNA-seq data from a variety of different model organisms allowed us to develop an RCP that can be used across all RNA-seq experiments hosted on GeneLab.
GL: What are the next steps?
EO and ASB: We are currently using the RCP to process RNA-seq data hosted on GeneLab, making DGE data publicly available for our users to further analyze and generate new hypotheses about the effects of the space environment on terrestrial biology. Since the field of bioinformatics is constantly changing and improving, we will revisit the GeneLab RCP annually and make any necessary updates, in collaboration with our AWG members, to ensure the pipeline stays up to date with the scientific community.

