Deep space exploration is the future of spaceflight, and with that comes many challenges in understanding how the human system is affected by the space environment to how long-term life support (ie food supply) can be sustained. A recent publication by Dr Manian, “Network Analysis of Gene Transcriptions of Arabidopsis thaliana in Spaceflight Microgravity,” explores one of these challenges by providing a fundamental knowledge of plant growth in spaceflight. Dr Vidya Manian, from the University of Puerto Rico, is a contributing member of the GeneLab analysis working group. In her recent publication, based on the reanalysis of two transcriptional profiling datasets, Dr Manian documents changes in Arabidopsis during spaceflight. GeneLab recently spoke to Dr Manian about this work, which was funded through a NASA EPSCoR call for the reuse of GeneLab data, and highlights how the GeneLab data systems and analysis working groups enabled this analysis.
GeneLab: What do you see as the most impactful result from this study?
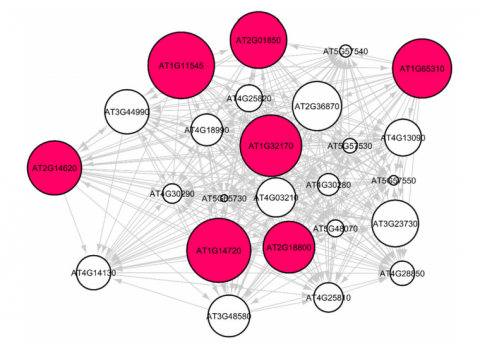
Vidya Manian: We showed that several novel algebraic graph theoretic methods in Gene-Regulatory Network analysis (Spectral methods, Hyper-linked Induced Topic Search (HITS) Algorithm, expansion, spectral gaps) can predict stress response of Arabidopsis thaliana in spaceflight microgravity environment. Our novel network analysis carries out genome to phenome level gene-regulatory predictions—by identifying key hub genes and authority genes that are involved in the stress responses. While finding hub genes that are differentially expressed under varying environmental conditions is useful, we also wanted to understand the influence of the environment on their regulatory pathways. The GLDS-120 is an interesting dataset that has given rise to important findings in previous literature as well as this one. Our rigorous network analysis showed that the phyD mutant of Arabidopsis adapted to the spaceflight total light environment with the lowest transcriptional cost, having lower values for several network measures. Our analysis identified genes that play a key role in the plant’s cell wall remodeling processes in spaceflight microgravity.
GL: How did the GeneLab Data Systems enable this work?
VM: The GeneLab data system was the primary resource of data for the study of Arabidopsis root growth in spaceflight. The latest RNA gene sequencing technology-based Arabidopsis transcriptomics datasets are available for the research community to download for free from their website. The publications of the PI’s and investigators of GLDS-7 and 120, posted on the GeneLab Data Systems have been helpful to us. Also, this work would not have been possible without the excellent measurement infrastructure provided by NASA in the International Space Station, and the expert personnel who carried out the experiments. The documents, guidelines, orientation and some of the tools such as TOAST provided by the GeneLab have been quite useful.
GL: How did the GeneLab Analysis Working Groups help you achieve your goals?
VM: The GeneLab AWG gave us useful information and feedback on our analysis. The AWG has provided visionary research infrastructure for us, including Webinars. This work was supported by a NASA EPSCoR grant. Dr. Jonathan Galazka is a collaborator in this grant. He was helpful in our initial formulation of the proposal that obtained the grant and provided support for the research. The NASA program monitors, Dr. Lester Morales and Dr. Egle Cekanaviciute have also guided us.
GL: What are your next steps?
VM: We are using (and will use) our novel techniques and some additional mathematical, computational, and algorithmic tools (machine learning approaches) to carry out analysis of the effects of microgravity environment stresses in spaceflight on plants, animals and humans.
By providing a portal linking processed data to flight parameters, GeneLab enables the exploration of molecular network responses of terrestrial biology to the space environment. With continued efforts to help the scientific community and the population at large gain new knowledge from space omics data, more insights can be gained into enabling deep space exploration.

